LIN-42, the Caenorhabditis elegans PERIOD homolog, Negatively
4.9 (218) · $ 18.99 · In stock
Author Summary MicroRNAs play pervasive roles in controlling gene expression throughout animal development. Given that individual microRNAs are predicted to regulate hundreds of mRNAs and that most mRNA transcripts are microRNA targets, it is essential that the expression levels of microRNAs be tightly regulated. With the goal of unveiling factors that regulate the expression of microRNAs that control developmental timing, we identified lin-42, the C. elegans homolog of the human and Drosophila period gene implicated in circadian gene regulation, as a negative regulator of microRNA expression. By analyzing the transcriptional expression patterns of representative microRNAs, we found that the transcription of many microRNAs is normally highly dynamic and coupled aspects of post-embryonic growth and behavior. We suggest that lin-42 functions to modulate the transcriptional output of temporally-regulated microRNAs and mRNAs in order to maintain optimal expression of these genes throughout development.

LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans
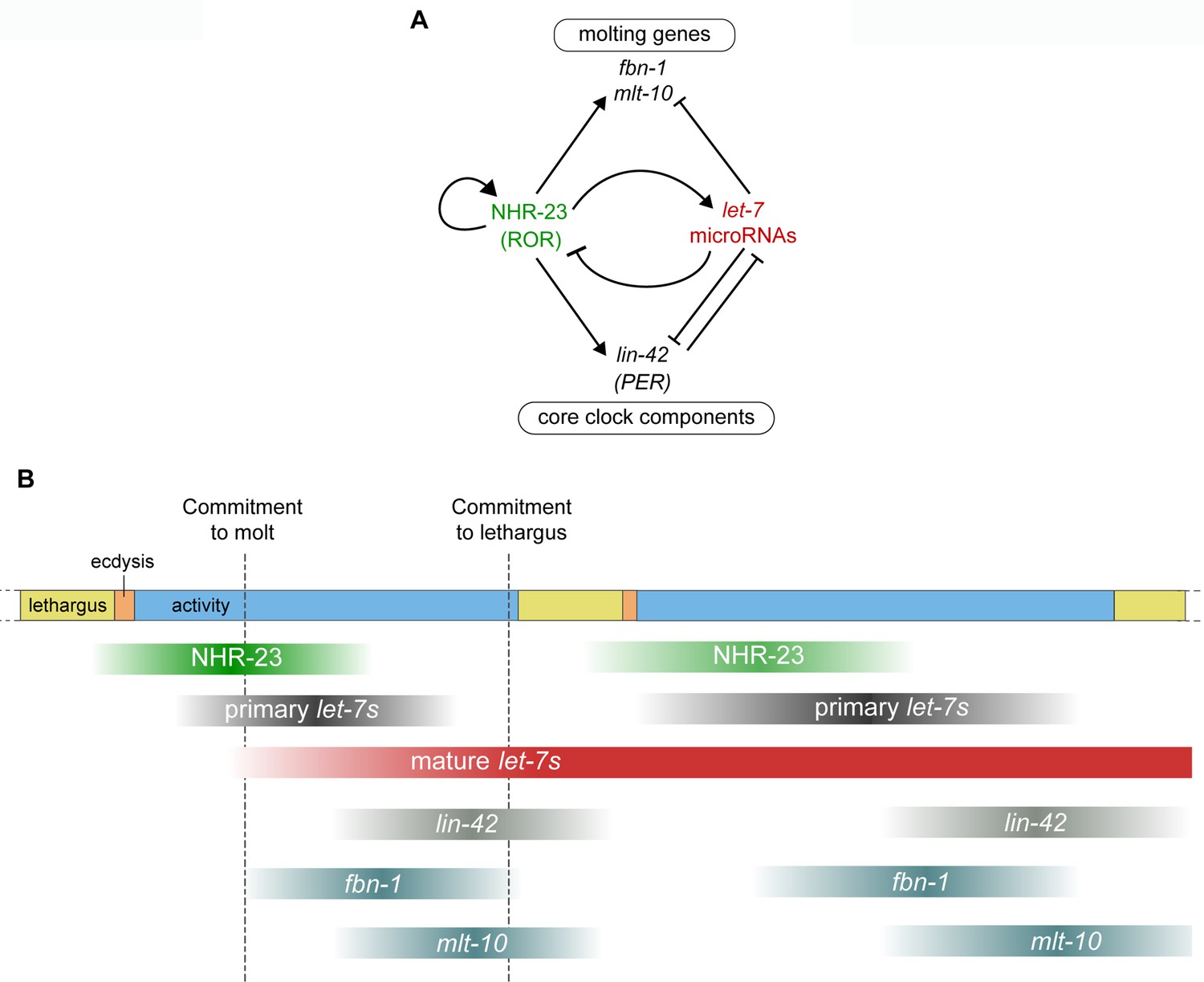
Feedback between a retinoid-related nuclear receptor and the let-7 microRNAs controls the pace and number of molting cycles in C. elegans
Dietary and microbiome factors determine longevity in Caenorhabditis elegans

JDB, Free Full-Text
Scientists Identify a Key Regulator of Developmental Timing

A circadian-like gene network programs the timing and dosage of heterochronic miRNA transcription during C. elegans development - ScienceDirect

DNA damage checkpoint and repair: From the budding yeast Saccharomyces cerevisiae to the pathogenic fungus Candida albicans - Computational and Structural Biotechnology Journal
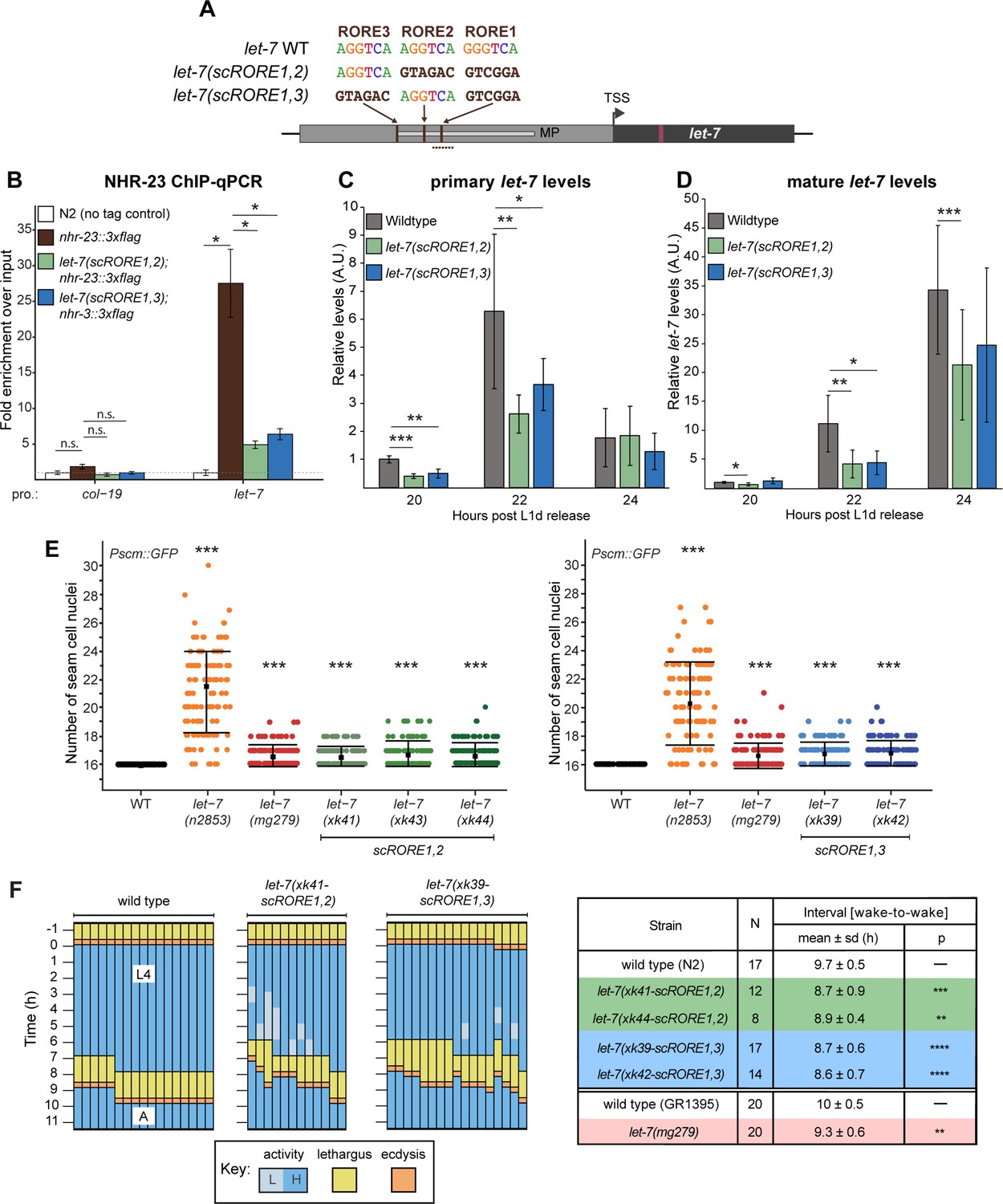
Feedback between a retinoid-related nuclear receptor and the let-7 microRNAs controls the pace and number of molting cycles in C. elegans
Age-related micro-RNA abundance in individual C. elegans